- Mo 11 September 2017
- MetaAnalysis
- Peter Schuhmacher
- #Python, #Statistics, #Effect size
We follow the following publication:
Ding-Geng (Din) Chen, Karl E. Peace (2013): Applied Meta-Analysis with R. CRC Press, Taylor & Francis Group, Boca Raton (FL), 321pp. ISBN-10: 1466505990, ISBN-13: 978-1466505995. See also here
The applications in this publication are programmed in R. We do it in Python here. We will describe the cases in a minimalistic way only. For details consult the book please.
The case
The example in chapter 1 mimics a multi center study of a diastolic blood pressure decreasing drug. The data are generated by ading random noise to a normal distribution. The generated data are concatenated to a data framework which allows for queries, statistical analysis and graphical displays. As a result we get a running workflow for a typical type of analysis.
The example generates for each of the 5 clinical centers the data series for the control group and for the treated group. A series includes the variables age, base pressure, endpoint pressure, difference between them, the number of the clinical center and the type of treament ('drug' = yes, 'ctrl' = no)
Implementation with Python
Import the Python librarys
The add-on libraries to the core of Python are NumPy for the matrix/array data types, and Pandas for data structures and prelimnary data analysis. The statistical package StatsModels is used for the regression model and the analysis of variances. Seaborn is used for the grafics which is based on Matplotlib
import numpy as np
import pandas as pd
import statsmodels.api as sm
import statsmodels.formula.api as smf
import seaborn as sns
sns.set(style="ticks")
%matplotlib inline
Define the functions to generate the data
The first 2 moments - mean and sdev - are used as input to generate a normal distributed data series.
def generateData(nobs,center,treatment,bp_μ, bp_σ,bp_δμ, age_μ, age_σ):
# nobs = n observations
# bp_μ = blood pressure mean
# bp_σ = blood pressure sdev
# bp_δμ= mean change of blood pressure due to drug; = 0 for no drug
# bp_Δ = difference between endpoint and baseline of the clinical trial
bp_base = np.random.normal(bp_μ , bp_σ, nobs) # random normal
bp_end = np.random.normal(bp_μ - bp_δμ, bp_σ, nobs) # random normal
bp_Δ = bp_end - bp_base
age = np.random.normal(age_μ, age_σ, nobs) # age, random normal
cen = center*np.ones_like(age,dtype=np.int) # number of the clinical center
trt = [treatment for t in range(nobs)] # treatment: with drug or ctrl
index = np.arange(nobs) # data index
myDataStack = {'bp_base': bp_base,'bp_end':bp_end, 'bp_Δ':bp_Δ,
'age':age,'cen':cen,'trt':trt}
return pd.DataFrame(myDataStack,index=index)
Set the input data
In this example it is assumed that 5 clinical centers are involved in the multi center study and that bp_δμ (change of mean blood pressure) is the only parameter that varies between the centers. For the control group bp_δμ_ctrl = 0
#------ main --------------------------------
#---- data we keep constant in this example:
nObs=100; # n observations
trt_ctrl = 'ctrl' # control group
trt_drug = 'drug' # group with treatment
bp_δμ_ctrl = 0 # change of mean blood pressure in the ctrl-group
bp_μ = 100 # mean blood pressure
bp_σ = 10 # sdev blood pressure
age_μ = 50 # mean age
age_σ = 10 # sdev age
#----- data we change to build the series
center1 = 1; bp_δμ_1 = 10 # clinical center; mean treatment effect
center2 = 2; bp_δμ_2 = 13
center3 = 3; bp_δμ_3 = 15
center4 = 4; bp_δμ_4 = 8
center5 = 5; bp_δμ_5 = 10
Now generate the data series and build a DataFrame
Each clinical center has a data series of the same length for the control group and the treated group. All data are concatenated into 1 data frame.
# series 1
dat4ctrl = generateData(nObs,center1,trt_ctrl,bp_μ,bp_σ,bp_δμ_ctrl,age_μ, age_σ)
dat4drug = generateData(nObs,center1,trt_drug,bp_μ,bp_σ,bp_δμ_1 ,age_μ, age_σ)
d1 = pd.concat([dat4ctrl,dat4drug],ignore_index=True)
# series 2
dat4ctrl = generateData(nObs,center2,trt_ctrl,bp_μ,bp_σ,bp_δμ_ctrl,age_μ, age_σ)
dat4drug = generateData(nObs,center2,trt_drug,bp_μ,bp_σ,bp_δμ_2 ,age_μ, age_σ)
d2 = pd.concat([dat4ctrl,dat4drug],ignore_index=True)
# series 3
dat4ctrl = generateData(nObs,center3,trt_ctrl,bp_μ,bp_σ,bp_δμ_ctrl,age_μ, age_σ)
dat4drug = generateData(nObs,center3,trt_drug,bp_μ,bp_σ,bp_δμ_3 ,age_μ, age_σ)
d3 = pd.concat([dat4ctrl,dat4drug],ignore_index=True)
# series 4
dat4ctrl = generateData(nObs,center4,trt_ctrl,bp_μ,bp_σ,bp_δμ_ctrl,age_μ, age_σ)
dat4drug = generateData(nObs,center4,trt_drug,bp_μ,bp_σ,bp_δμ_4 ,age_μ, age_σ)
d4 = pd.concat([dat4ctrl,dat4drug],ignore_index=True)
# series 5
dat4ctrl = generateData(nObs,center5,trt_ctrl,bp_μ,bp_σ,bp_δμ_ctrl,age_μ, age_σ)
dat4drug = generateData(nObs,center5,trt_drug,bp_μ,bp_σ,bp_δμ_5 ,age_μ, age_σ)
d5 = pd.concat([dat4ctrl,dat4drug],ignore_index=True)
# build 1 DataFrame with all data
dAll = pd.concat([d1,d2,d3,d4,d5],ignore_index=True)
Let's have a look at the head of the first few lines of the data frame:
dAll.head(5)
| age | bp_base | bp_end | bp_Δ | cen | trt | |
|---|---|---|---|---|---|---|
| 0 | 60.720858 | 111.664456 | 99.437909 | -12.226547 | 1 | ctrl |
| 1 | 36.035688 | 89.109796 | 88.445715 | -0.664080 | 1 | ctrl |
| 2 | 38.823806 | 92.925221 | 97.549218 | 4.623998 | 1 | ctrl |
| 3 | 38.783952 | 91.520139 | 104.185235 | 12.665097 | 1 | ctrl |
| 4 | 65.327808 | 93.283620 | 104.308016 | 11.024396 | 1 | ctrl |
Print the statistics of the data
dAll.groupby(['cen', 'trt']).agg(['count','mean','std'])
| age | bp_base | bp_end | bp_Δ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | mean | std | count | mean | std | count | mean | std | count | mean | std | ||
| cen | trt | ||||||||||||
| 1 | ctrl | 100 | 48.467260 | 10.194531 | 100 | 98.567163 | 11.792550 | 100 | 99.793300 | 9.211106 | 100 | 1.226136 | 13.389781 |
| drug | 100 | 49.628733 | 9.628830 | 100 | 99.935044 | 10.373236 | 100 | 87.838175 | 9.571521 | 100 | -12.096870 | 14.092291 | |
| 2 | ctrl | 100 | 49.215135 | 10.550840 | 100 | 98.480676 | 10.410645 | 100 | 99.435871 | 10.324950 | 100 | 0.955195 | 14.974590 |
| drug | 100 | 50.154245 | 10.313113 | 100 | 98.144471 | 9.870396 | 100 | 86.472468 | 10.531072 | 100 | -11.672004 | 14.280153 | |
| 3 | ctrl | 100 | 47.850356 | 9.693100 | 100 | 99.867286 | 9.946296 | 100 | 99.712728 | 9.894162 | 100 | -0.154558 | 14.311047 |
| drug | 100 | 49.427241 | 10.435070 | 100 | 98.287170 | 10.428239 | 100 | 86.059913 | 10.143901 | 100 | -12.227257 | 14.219824 | |
| 4 | ctrl | 100 | 48.263132 | 9.648512 | 100 | 99.514863 | 9.863520 | 100 | 99.043574 | 9.933129 | 100 | -0.471289 | 13.328751 |
| drug | 100 | 49.930536 | 10.266096 | 100 | 99.877485 | 10.086439 | 100 | 91.581340 | 9.771715 | 100 | -8.296145 | 13.936570 | |
| 5 | ctrl | 100 | 50.424546 | 9.768934 | 100 | 99.979968 | 10.454416 | 100 | 100.016502 | 10.191461 | 100 | 0.036535 | 14.902816 |
| drug | 100 | 49.665251 | 10.003291 | 100 | 98.986446 | 9.906621 | 100 | 90.535263 | 9.178024 | 100 | -8.451183 | 13.524656 | |
Play around with the different types of plots
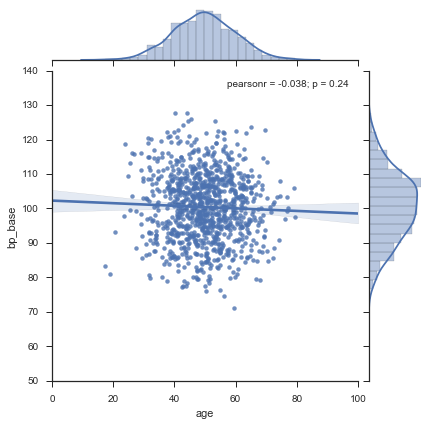
This first plot contains all data, age vs base blood pressure, treated and untreated over all 5 clinical centers. On the east and the north side of the graph you should recognize the generated normal distributions in the bar plots, whereas the plotted lines are nice looking splines over the bar data only. The plotted line in the graph with y = base blood pressure should be horizontal most of the time and have a value of about 100 beceause this the mean given as input.
Each data series has 100 observations, so 1'000 data points are included in this graph.
sns.jointplot(x="age", y="bp_base", data= dAll, kind="reg");
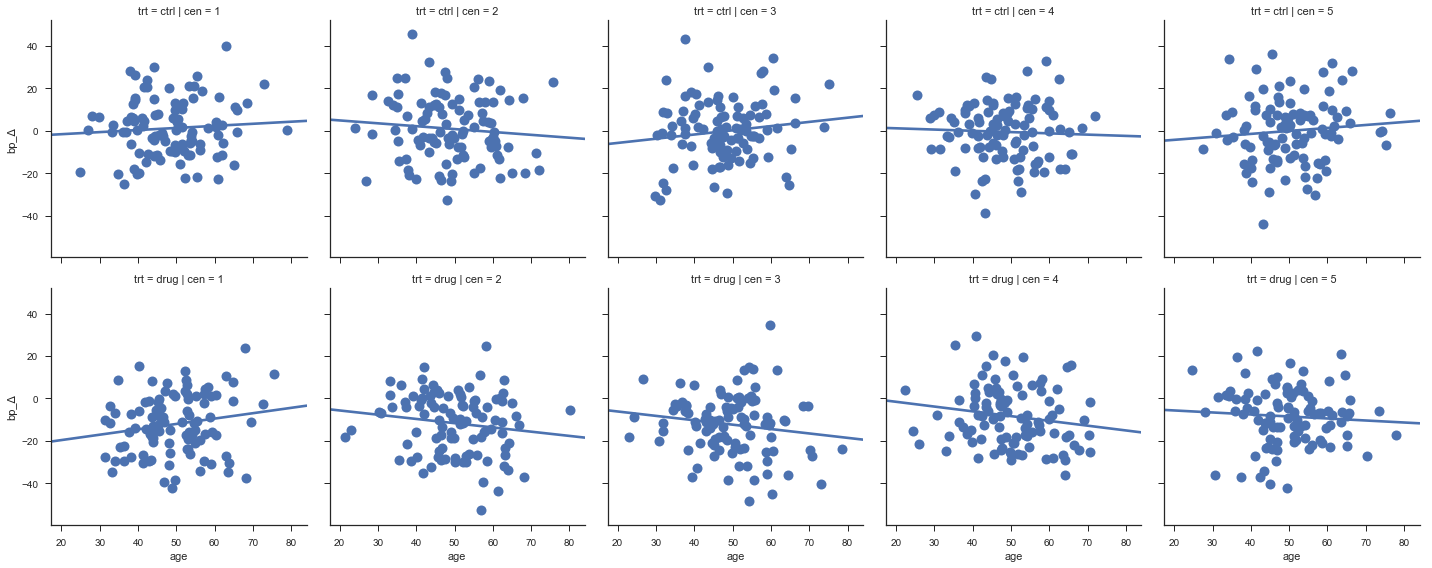
The next graph displays the change of blood pressure vs age for each center. The change of blood pressure should be more negative for the treated groups than for the control group hopefully, beceause this corresponds to the mean given as input.
sns.lmplot(x="age", y="bp_Δ", col="cen", row="trt", data=dAll,
ci=None, palette="muted", size=4,
scatter_kws={"s": 100, "alpha": 1 });
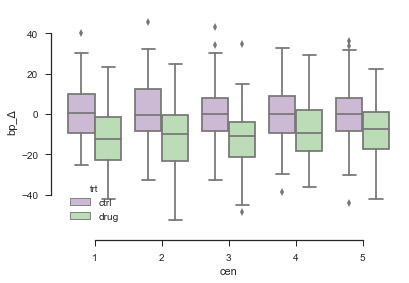
The change of blood pressure, displayed for the ctrl-group and the trt-group for each clinical center, is summarized in a box plot.
sns.boxplot(x="cen", y="bp_Δ", hue="trt", data=dAll, palette="PRGn")
sns.despine(offset=10, trim=True)
Data analysis of each center
With these preliminary graphical illustrations, we now proceed to data analysis. We start with the individual center effects. We subset the data from each center to perform an analysis of variance. We consider a treament effect as statistically significant at p-value \(<0.05\) e.g.
e_c1 = smf.ols(formula='bp_Δ ~ trt', data = dAll[dAll["cen"]==1]).fit()
t_c1 = sm.stats.anova_lm(e_c1, typ=1)
print('Center = 1'); print(t_c1, '\n')
e_c2 = smf.ols(formula='bp_Δ ~ trt', data = dAll[dAll["cen"]==2]).fit()
t_c2 = sm.stats.anova_lm(e_c2, typ=1)
print("Center = 2"); print(t_c2, '\n')
e_c3 = smf.ols(formula='bp_Δ ~ trt', data = dAll[dAll["cen"]==3]).fit()
t_c3 = sm.stats.anova_lm(e_c3, typ=1)
print("Center = 3"); print(t_c3, '\n')
e_c4 = smf.ols(formula='bp_Δ ~ trt', data = dAll[dAll["cen"]==4]).fit()
t_c4 = sm.stats.anova_lm(e_c4, typ=1)
print("Center = 4"); print(t_c4, '\n')
e_c5 = smf.ols(formula='bp_Δ ~ trt', data = dAll[dAll["cen"]==5]).fit()
t_c5 = sm.stats.anova_lm(e_c5, typ=1)
print("Center = 5"); print(t_c5, '\n')
Center = 1
df sum_sq mean_sq F PR(>F)
trt 1.0 8875.124407 8875.124407 46.97338 8.913676e-11
Residual 198.0 37410.010029 188.939445 NaN NaN
Center = 2
df sum_sq mean_sq F PR(>F)
trt 1.0 7972.307055 7972.307055 37.239755 5.404231e-09
Residual 198.0 42387.948440 214.080548 NaN NaN
Center = 3
df sum_sq mean_sq F PR(>F)
trt 1.0 7287.503584 7287.503584 35.809995 1.003735e-08
Residual 198.0 40293.937324 203.504734 NaN NaN
Center = 4
df sum_sq mean_sq F PR(>F)
trt 1.0 3061.418522 3061.418522 16.464392 0.000071
Residual 198.0 36816.473763 185.941787 NaN NaN
Center = 5
df sum_sq mean_sq F PR(>F)
trt 1.0 3602.067373 3602.067373 17.787537 0.000038
Residual 198.0 40096.015373 202.505128 NaN NaN
Data Analysis with pooled data from five centers
Remember data generation: Different treatment effects has been assigned to the different clinical centers but treatment has not been related to age. We hope to recognize that in the analysis again of course. We start to fit a model for the blood pressure difference bp_Δ with 3-way interactions among treatment, center and age including the covariates as follows:
We perform an analysis of variance on that model and reduce the complexity of the model afterwards motivated by the p-values as a ranking parameter. This gives the following series of models
Since the increase of the squared sums of the residuals (ssr) is moderate we may conclud that the simplest model is still accurate.
e_pool1 = smf.ols(formula='bp_Δ ~ trt*cen*age', data = dAll).fit()
t_pool1 = sm.stats.anova_lm(e_pool1, typ=1)
print("Pool = all");
print(t_pool1.sort_values("PR(>F)",axis=0)) # output table sorted by PR(>F)
Pool = all
df sum_sq mean_sq F PR(>F)
trt 1.0 29523.440890 29523.440890 149.114727 4.754130e-32
trt:cen 1.0 1047.327006 1047.327006 5.289759 2.165806e-02
trt:age 1.0 555.765934 555.765934 2.807020 9.416772e-02
trt:cen:age 1.0 370.249746 370.249746 1.870029 1.717815e-01
cen 1.0 235.404079 235.404079 1.188961 2.758041e-01
cen:age 1.0 111.981001 111.981001 0.565585 4.521971e-01
age 1.0 86.740679 86.740679 0.438103 5.081931e-01
Residual 992.0 196407.518177 197.991450 NaN NaN
e_pool2 = smf.ols(formula='bp_Δ ~ cen + trt*age', data = dAll).fit()
e_pool3 = smf.ols(formula='bp_Δ ~ cen + trt + age', data = dAll).fit()
e_pool4 = smf.ols(formula='bp_Δ ~ cen + trt ', data = dAll).fit()
table = sm.stats.anova_lm(e_pool1, e_pool2, e_pool3, e_pool4, typ=1)
print(table)
df_resid ssr df_diff ss_diff F Pr(>F)
0 992.0 196407.518177 0.0 NaN NaN NaN
1 995.0 197953.658419 -3.0 -1546.140243 2.587547 NaN
2 996.0 198479.145133 -1.0 -525.486714 2.638289 NaN
3 997.0 198579.582543 -1.0 -100.437410 0.504262 NaN
