- So 25 März 2018
- MetaAnalysis
- Peter Schuhmacher
- #numerical, #statistics, #python, #markov model
import numpy as np
import pandas as pd
from scipy import stats
from matplotlib import pyplot as plt
import matplotlib.colors as mclr
Why a change could be indicated
The decision tree is a simple form of decision model. But there are also limitions of the decision tree evident:
- a formal aspect is that the tree format becomes rapidly unwiedly when a combination of several options have to be mapped
- with regard to content the elapse of time is not explicit in decision trees. Many chronic diseases such as diabetes, ischaemic heart disease or some forms of cancer have a recuring-remitting pattern over a period of many years. If a longer time horizon has to be adopted, several features may become necessary to be modelled:
- continuing risk of recurrence
- competing risk of death as the cohort ages
- other clinical developments
The Markov model is an approach to handel added modelling options. The key structure of a markov model is:
- it is structured around disease states
- it is driven by a set of possible transitions between the disease states
- it can be run over a series of time periods which gives an insight over the temporal evolution of the disease states
- costs may be included in parallel
- the modelled transistions probabilities may change over time too, so that changing conditions may be included
We give in the follwing a basic example of a Markov model that illustrates the temporal evolution of a communicable disease in a population.
$$
\begin{equation}
\begin{array}{rcl}
\textrm{Markov matrix} \; \mathbf{M} &=& \left[\begin{matrix}0.721 & 0.202 & 0.067 & 0.010 \\
0.000 & 0.581 & 0.407 & 0.012 \\
0.000 & 0.000 & 0.750 & 0.250 \\
0.000 & 0.000 & 0.000 & 1.000 \end{matrix}\right] \\
\textrm{Start vector} \;\; \mathbf{p}_0 &=& \left[\begin{matrix}1 \\ 0 \\0 \\0\end{matrix}\right]\\
Repeat:\\
\textrm{Time step} \;\; \mathbf{p}_1 &=& \mathbf{M}^T \cdot \mathbf{p}_0 \\
\textrm{Iteration} \;\; \mathbf{p}_0 &:=& \mathbf{p}_1
\end{array}
\end{equation}
$$
d1
Set up the Markov system
M_states = np.array(['A (healthy)', 'B (infected)', 'C (ill)', 'D (dead)'])
MM = np.array([[0.721, 0.202, 0.067, 0.010],
[0.000, 0.581, 0.407, 0.012],
[0.000, 0.000, 0.750, 0.250],
[0.000, 0.000, 0.000, 1.000] ])
dmm = pd.DataFrame(MM, columns=M_states, index=M_states); dmm
| A (healthy) | B (infected) | C (ill) | D (dead) | |
|---|---|---|---|---|
| A (healthy) | 0.721 | 0.202 | 0.067 | 0.010 |
| B (infected) | 0.000 | 0.581 | 0.407 | 0.012 |
| C (ill) | 0.000 | 0.000 | 0.750 | 0.250 |
| D (dead) | 0.000 | 0.000 | 0.000 | 1.000 |
Run the Markov simulation
We run the simulation with a population of 1000 members. Note that simulation part has 7 lines of code only:
nRuns = 21
m_result = np.zeros((nRuns,len(M_states)))
v0 = np.array([1, 0, 0, 0])*1000
for ir in range(nRuns):
m_result[ir,:] = v0
v0 = np.dot(MM.T,v0)
dmr = pd.DataFrame(np.rint(m_result).astype(int), columns=M_states)
Plot the result
plot_gr02(dmr,'time','# of people','Markov simulation of a communicable disease'); dmr
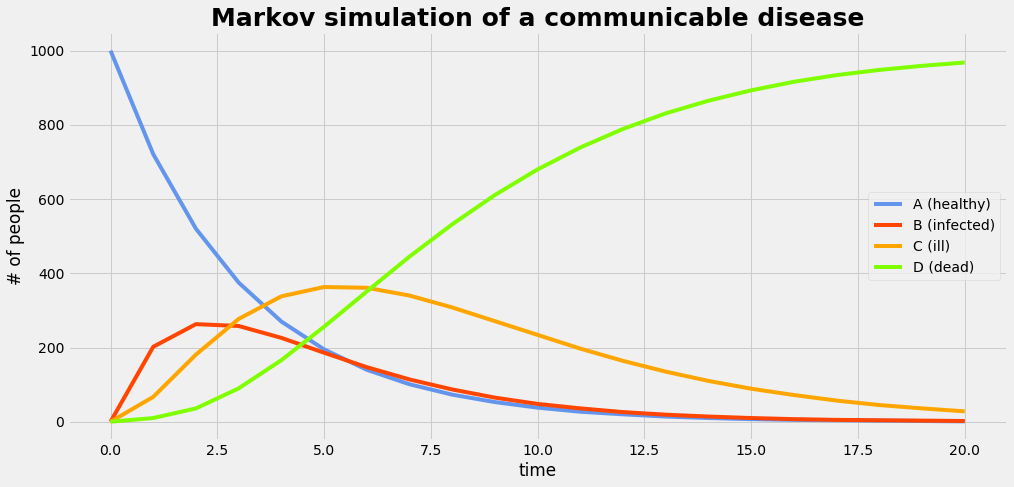
| A (healthy) | B (infected) | C (ill) | D (dead) | |
|---|---|---|---|---|
| 0 | 1000 | 0 | 0 | 0 |
| 1 | 721 | 202 | 67 | 10 |
| 2 | 520 | 263 | 181 | 36 |
| 3 | 375 | 258 | 277 | 90 |
| 4 | 270 | 226 | 338 | 166 |
| 5 | 195 | 186 | 363 | 256 |
| 6 | 140 | 147 | 361 | 351 |
| 7 | 101 | 114 | 340 | 445 |
| 8 | 73 | 87 | 308 | 532 |
| 9 | 53 | 65 | 271 | 611 |
| 10 | 38 | 48 | 234 | 680 |
| 11 | 27 | 36 | 197 | 739 |
| 12 | 20 | 26 | 164 | 789 |
| 13 | 14 | 19 | 135 | 831 |
| 14 | 10 | 14 | 110 | 865 |
| 15 | 7 | 10 | 89 | 893 |
| 16 | 5 | 7 | 72 | 916 |
| 17 | 4 | 5 | 57 | 934 |
| 18 | 3 | 4 | 45 | 948 |
| 19 | 2 | 3 | 36 | 959 |
| 20 | 1 | 2 | 28 | 968 |
import graphviz as gv
def plotMarkovModel():
d1 = gv.Digraph(format='png',engine='dot')
c = ['cornflowerblue','orangered','orange','chartreuse']
d1.node('A','A (healthy)', style='filled', color=c[0])
d1.node('B','B (infected)',style='filled', color=c[1])
d1.node('C','C (ill)', style='filled', color=c[2])
d1.node('D','D (dead)', style='filled', color=c[3])
d1.edge('A','A',label='0.721'); d1.edge('A','B',label='0.202'); d1.edge('A','C',label='0.067'); d1.edge('A','D',label='0.010')
d1.edge('B','B',label='0.581'); d1.edge('B','C',label='0.407'); d1.edge('B','D',label='0.012')
d1.edge('C','C',label='0.750'); d1.edge('C','D',label='0.250')
d1.edge('D','D',label='1.000');
return d1
d1 = plotMarkovModel()
d1.render('img/mamo1', view=True)
def plot_gr02(DF,xLabel,yLabel,grTitel):
with plt.style.context('fivethirtyeight'):
fig = plt.figure(figsize=(15,7)) ;
ax1 = fig.add_subplot(111);
#DF.plot(ax = plt.gca())
colors = ['cornflowerblue','orangered','orange','chartreuse']
DF.plot(ax = ax1, style='-', color=colors)
plt.xlabel(xLabel); plt.ylabel(yLabel);
plt.title(grTitel, fontsize=25, fontweight='bold');
plt.show()
